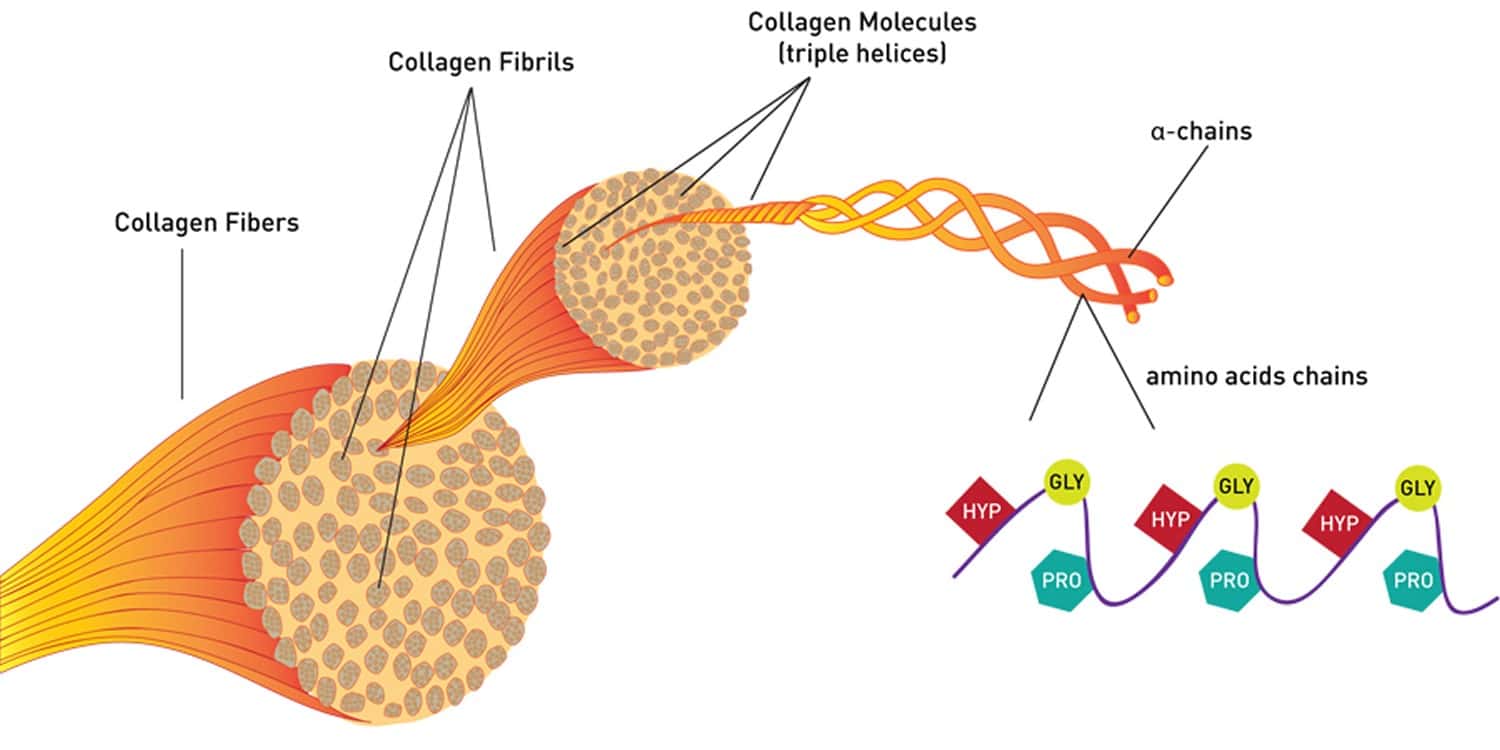
The importance of measuring Clostridium histolyticum collagenases’ collagen degradation activity is illustrated by the “enzyme problem” that arose before beginning an NIH-funded, multi-million dollar clinical trial to assess the clinical efficacy of human islet transplantation to manage adult type 1 diabetic patients.
C. histolyticum collagenase has been used since the 1960s as the primary enzyme for enzyme-mediated cell isolation methods. It was first purified in 1953 (1). In 1995, Boehringer Mannheim Biochemicals introduced the first commercially purified collagenase-protease enzyme mixture designed to improve islet recovery from human pancreas.(2) The inability of human islet isolators to recover sufficient islets for successful islet transplants in adult diabetic patients stimulated the development of Liberase™ HI Purified Enzyme Blend product.
Liberase HI became the “gold standard” enzyme for isolating human islets for > 10 years.(3) However, changes in the enzyme purification process ≈ 2002-2004 led to the manufacture of inconsistent Liberase HI products, leading to more failed islet isolations where islet recovery was insufficient for subsequent transplantation.(4) In 2007, NIH discontinued the planned use of Liberase HI in their Clinical Islet Transplantation Consortium (CITC) clinical trials for allo-islet transplantation. NIH forbade the use of Liberase HI because of the potential threat of transmission of spongiform encephalopathy from bovine proteins used in the C. histolyticum fermentation culture media.(3) Nordmark’s NB-1 Collagenase and NB Protease products were used instead of Liberase HI to isolate islets used in the Consortium’s clinical trial.
Many centers adopted Nordmark products, but this switch caused concern for three of the most experienced islet transplant centers since they could not obtain comparable islet yields to those obtained with Liberase HI.(5) Biochemical and enzyme activity analysis at VitaCyte led to one explanation: Nordmark’s NB-1 collagenase contained reduced collagen degradation activity (CDA) compared to Roche HI and VitaCyte’s Collagenase HA products. Moreover, a comparison of human islet isolation results performed using VitaCyte’s Collagenase HA-thermolysin enzyme mixtures to those using Nordmark’s NB-1 Collagenase-NB Protease mixtures showed that Collagenase HA gave significantly shorter digestion times and had significantly higher human islet yields as measured by purified IEQ (islet equivalent number) per organ or g of trimmed tissue.(6)
Nordmark’s and VitaCyte’s collagenase products used similar doses of Wunsch activity for islet isolation, the accepted standard for measuring collagenase activity, but this assay did not predict the isolation results. Why?
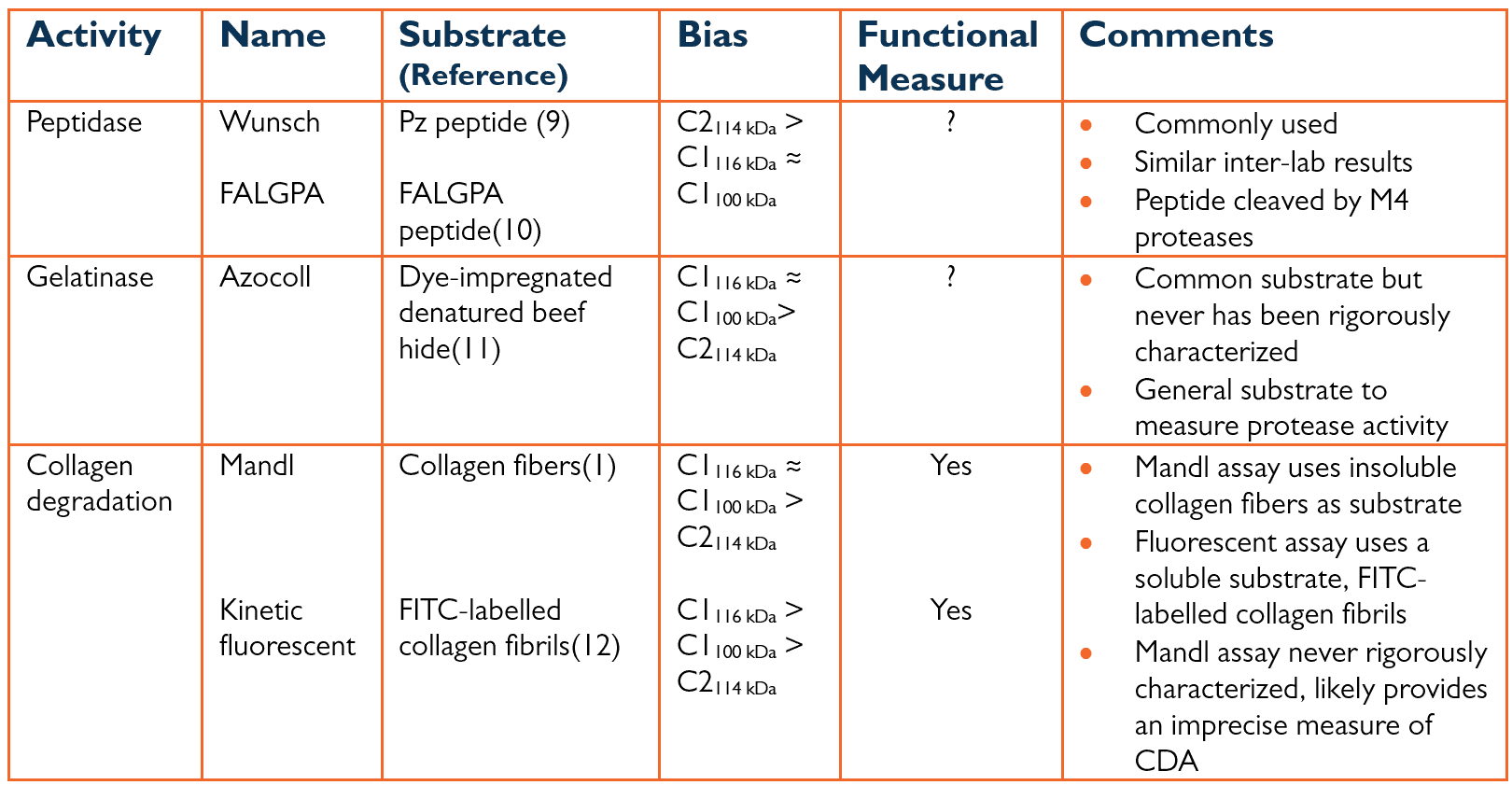
The initial purification and characterization of C. histolyticum collagenase showed that two classes of collagenase, C1 and C2 collagenase, worked synergistically to degrade native collagen.(7, 8) The three different assays commonly used to measure collagenase activity are summarized in Table 1 below. Class I (C1) collagenase had a higher specific gelatinase activity (U/mg protein) than class II (C2) collagenase. The converse was true for C2, which had much higher specific activities for peptide substrates than C1. There was no knowledge of how these two classes degraded native collagen.
Table 1: Assays Used to Measure Collagenase Activity
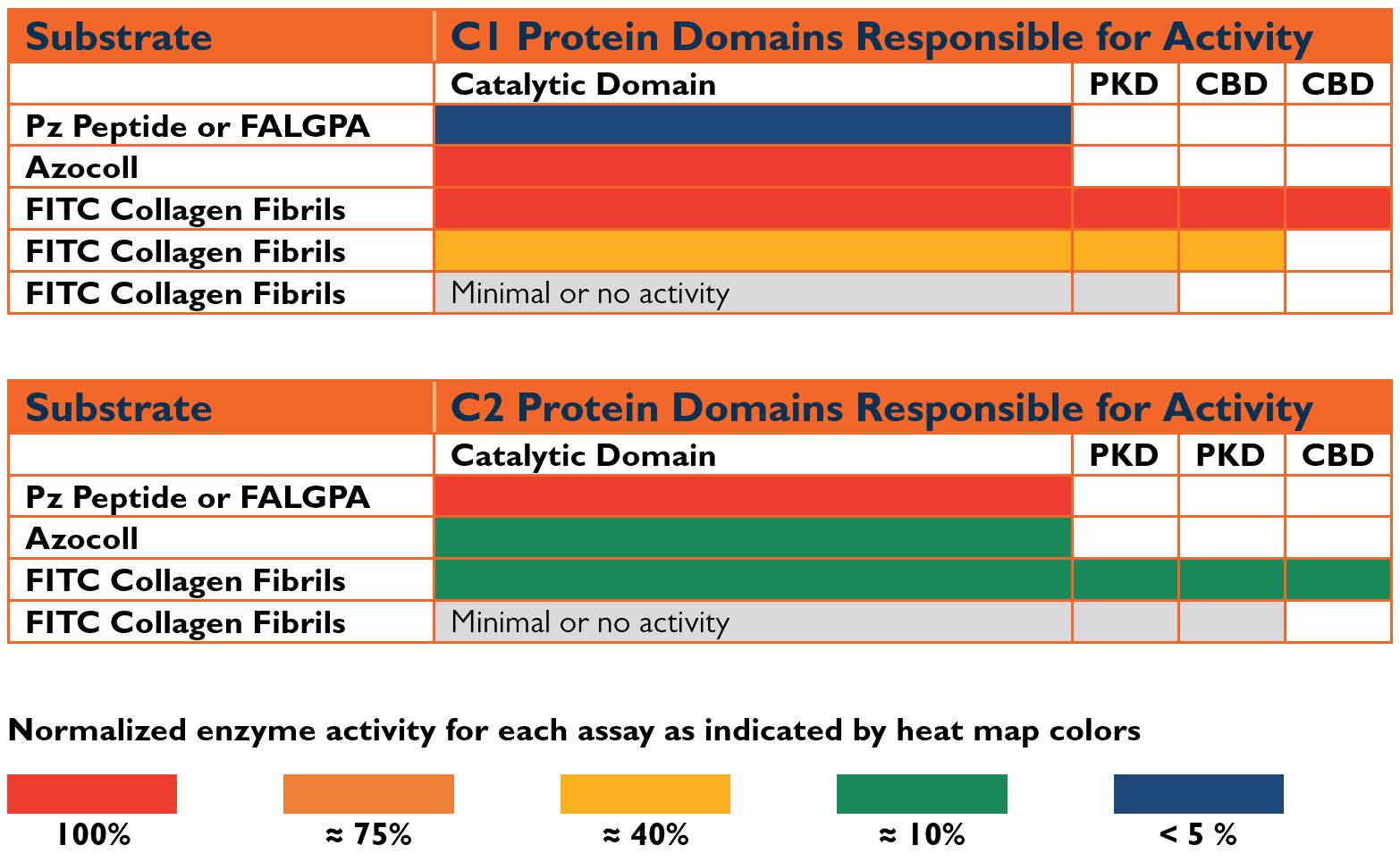
The C. histolyticum C1 and C2 genes were sequenced and subsequently expressed as recombinant enzymes by Matsushita’s laboratory.(13) C1 and C2 collagenases contained three functional domains: a catalytic domain that degraded/cut the collagen, a linking or PKD domain with a protein sequence similar to a protein found in human polycystic kidney disease, and a collagen-binding domain (CBD). Intact C1 (C1116 kDa) has a catalytic domain, a PKD domain, and two collagen binding domains, whereas intact C2 (C2114 kDa)has a catalytic domain, 2 PKD domains, and a single CBD. The catalytic domain accounts for about 66% of the intact enzymes.
Effective collagen degradation requires the C1 and C2 collagenase enzymes to contain a catalytic domain and at least one CBD. (14) There are three functional forms of collagenase where the function is defined by the ability to degrade collagen: C1116 kDa, C2114 kDa, and a truncated form of C1 (C1100 kDa). Proteolysis of C1’s carboxy-terminal CBD during the fermentation or purification process creates the C1100 kDa form. This form can degrade collagen since it contains one CBD.
Figure 1 below correlates gene structure to enzyme activity using different “heat map” colors to indicate the minimal structure required to measure active enzymes. The Figure shows a major limitation of Wunsch activity since it measures C2, not C1 activity. Moreover, peptidase and gelatinase activities only require the catalytic domain for activity. Consequently, gelatinase or peptidase activity measurement is ambiguous since it does not differentiate between functional and non-functional collagenases.
Figure 1: Identification of Minimal Protein Domain Structure Required for Collagenase Activity
By contrast, CDA is the only accurate measure of collagenase function, which is the degradation of native collagen. The extracellular matrix, rich in collagen and other proteins, usually is resistant to proteolytic degradation. Teleologically, collagen is likely responsible for this resistance. Collagen is highly conserved, indicating it is essential for survival. Mammalian collagenases are used for tissue remodeling and have narrow specificity for specific types of collagen.(15) In contrast, bacterial collagenases digest essentially all types of collagen, which is why it is used for many life science research and biomedical applications. Bacteria use collagenase to break down dead organic matter, creating peptides that the organism uses as foodstuff.
Structure-function studies using purified recombinant C. histolyticum C1 and C2 collagenases showed that each class has a different degradation mechanism. C1 collagenase is a processive protease that “shaves off” tropocollagen molecules from a collagen fibril or fiber. In contrast, C2 collagenase is an endoprotease that creates collagen polypeptides.(16) These two enzymes likely complement each other, with C2 creating breaks in the tropocollagen that enable C1 to bind and shave off each tropocollagen molecule.
The two primary assays used to measure CDA for commercial collagenase products are the Mandl assay (1) and a kinetic fluorescent microtiter plate CDA.(12) Mandl et al. developed a collagenase degradation assay in 1953 to detect collagenase activity during the initial purification of C. histolyticum collagenase. The Mandl assay is a research assay that uses insoluble collagen fibers as a substrate. Today, many commercial collagenase suppliers still use this assay to measure CDA in their commercial collagenase products. It has never been rigorously characterized.
Recently, VitaCyte internally developed a modified Mandl assay that ensured the assay conditions followed pseudo-first-order kinetics, generating a linear dose-response curve. This analysis showed little difference between the specific CDAs of the C1116 kDa and C1100 kDa forms. These specific activities were about four-fold higher than the specific activity of the C2114 kDa form.(16)
In the kinetic fluorescent CDA, collagenase unwinds and degrades soluble fluorescein isothiocyanate conjugated to bovine type I collagen fibrils (FITC-fibrils).(12, 16) Degradation is detected by an increase in FITC fluorescence over the assay period. Fluorescence increases because the FITC protein conjugate is relaxed or freed from the collagen, overcoming the fluorescent self-quenching that occurs when the FITC is bound to the collagen fibrils. VitaCyte uses the kinetic fluorescent assay to report CDA values on the Certificate of Analyses for VitaCyte’s products. The between-run coefficient of variation for this assay is ≤ 15%. The CDA specific activity for the different forms of collagenase are summarized in Table 2 below.
Table 2: CDA Specific Activities of the 3 Functional Forms of Collagenase Enzymes

Resolution of the “enzyme problem:” In 2008, NIH approved Roche and VitaCyte as alternative suppliers of enzymes for the clinical trial. Roche developed a new Liberase, a Liberase MTF kit, that contained two bottles of natural purified collagenase (≈ 500 mg per bottle) and three bottles of Thermolysin (15 mg per bottle). The results from eight manufacturing islet manufacturing centers that used either Nordmark, Roche, or VitaCyte enzymes for isolating human islets for use in the CITC’s CIT-07 clinical trial showed that only 52% of isolations generated sufficient numbers of islets for subsequent transplant.(17) These results do not indicate the enzymes used for isolating the islets.
Take home message: if you want tighter control over your cell isolation procedure, consider purchasing collagenase enzymes from suppliers that include CDA results on their product’s certificate of analysis. If CDA values are reported, request more details on the assay procedure and characteristics. At a minimum, the supplier should provide information on the specific activities of collagenase and trypsin (or any other strong protease) detected by this assay. The use of trypsin (or any other general neutral protease) in the assay is used to ensure the specificity of the collagen degradation. If a neutral protease is active in the assay, it strongly indicates that native collagen substrate has been denatured and the assay measures gelatinase activity. The supplier should also be able to estimate the between-run variability, as determined by running a stable internal control standard in each assay.
Every new lot of FITC fibril substrate prepared for VitaCyte’s kinetic, fluorescent CDA assay is assessed for resistance to protease activity. These substrates are always negative for protease activity. Moreover, the between-run CV is about 15%.
If you have any questions about use of CDA assays to characterize C. histolyticum collagenase products, do not hesitate to contact us.
References
1. Mandl I, MacLennan JD, Howes EL. Isolation and characterization of proteinase and collagenase from Cl. histolyticum. J Clin Inv. 1953;32:1323-9.
2. Fetterhoff TJ, Cavanagh TJ, Wile KJ, Wright MJ, Dwulet FE, Gill J, et al. Human pancreatic dissociation using a purified enzyme blend. Transplant Proc. 1995;27:3282-3.
3. Wang Y, Paushter D, Wang S, Barbaro B, Harvat T, Danielson KK, et al. Highly purified versus filtered crude collagenase: comparable human islet isolation outcomes. Cell Transplant. 2011;20:1817-25.
4. Barnett MJ, Zhai X, LeGatt DF, Cheng SB, Shapiro AMJ, Lakey JRT. Quantitative assessment of collagenase blends for human islet isolation. Transplant. 2005;80:723-8.
5. McCarthy RC, Green ML, Dwulet FE. Evolution of enzyme requirements for human islet isolation. OBM Transplantation. 2018; 2:[1-30 pp.]. Available from: http://www.lidsen.com/journals/transplantation/transplantation-02-04-024.
6. Balamurugan AN, Breite AG, Anazawa T, Loganathan G, Wilhelm JJ, Papas KK, et al. Successful human islet isolation and transplantation indicating the importance of class 1 collagenase and collagen degradation activity assay. Transplantation. 2010 2010;89(8):954-61.
7. Kono T. Purification and partial characterization of collagenolytic enzymes from Clostridium histolyticum. Biochem. 1968;7:1106-14.
8. Mandl I, Keller S, Manahan J. Multiplicity of Clostridium histolyticum collagenases. Biochem. 1964;3:1737-41.
9. Wünsch E, Heidrich H-G. Zur quantitativen bestimmung der kollagenase. Hoppe-Seyler’s Zeitschrift Physiologische Chemie. 1963;333:149-51.
10. Van Wart HE, Steinbrink DR. A continuous spectrophotometric assay for Clostridium histolyticum collagenase. Anal Biochem. 1981;113:356-65.
11. Chavira R, Burnett TJ, Hageman JH. Assaying proteases with azocoll. Anal Biochem. 1984;136:446-50.
12. McCarthy RC, Spurlin B, Wright MJ, Breite AG, Sturdevant LK, Dwulet CS, Dwulet FE. Development and characterization of a collagen degradation assay to assess purified collagenase used in islet isolation. Transplant Proc. 2008;40:339-42.
13. Matsushita O, Okabe A. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon. 2001;39:1769-80.
14. Matsushita O, Jung CM, Katayama S, Minami J, Takahashi Y, Okabe A. Gene duplication and multiplicity of collagenases in Clostridium histolyticum. J Bacteriology. 1999;181:923-33.
15. Dioszegi M, Cannon P, Van Wart HE. Vertebrate collagenases. Meth Enzymol. 1995;248:413-31.
16. McCarthy RC, Sakon J. Webinar: Collagen degradation by Clostridal histolyticum collagenases 2021 [Available from: Collagen Degradation by Clostridial histolyticum Collagenases – VitaCyte (cloudwaysapps.com).
17. Ricordi C, Goldstein JS, Balamurugan AN, Szot GL, Kin T, Liu C, et al. National Institutes of Health-Sponsored Clinical Islet Transplantation Consortium Phase 3 Trial: Manufacture of a Complex Cellular Product at Eight Processing Facilities. Diabetes. 2016;65:3418-28.
